Genetic biodiversity is the fuel that drives crop improvement. However, in major cereal crops, while the traditional method of crop improvement has brought significant yield improvements over time, it has also resulted in a drastic loss of crop genetic biodiversity.
As a result, many of our crops have lost resilience, becoming potentially vulnerable to a range of stresses, from environmental factors to disease. Continued loss of genetic diversity could severely affect crop yields and threaten the sustainability of economically and socially important end uses.
So what’s the answer? Strategies for mobilising novel diversity into crop gene pools already exist, but none are currently effective for introducing the resilience afforded by traits conferred by many genes of small effect. Such traits include robust and durable tolerance to pests and pathogens, resilience to heat or drought, or basic environmental components driving yield, such as day length or altitude.
This project, co-funded by Scottish Government through the Rural and Environment Science and Analytical Services (RESAS) Division and UKRI, is investigating the merits of a radically different approach to refreshing the diversity in the genetically eroded barley breeding gene pool. We are evaluating a novel improvement strategy that involves speeding up the process of harnessing traits from wild barley genotypes and incorporating them into the genome of commercial varieties required for NW European agriculture. In taking this approach, we address a fundamental issue that has perplexed both the genetic resources and breeding communities for decades: how do we efficiently extract value from biodiverse wild germplasm in crop improvement programs, especially for complex and multigenic traits.
Photo: Field of barley (Credit: Joanne Russell)

Stage
Directory of Expertise
Purpose
The term ‘biodiversity loss’ is commonly associated with a threat to the loss of plant, animal, insect or microbial species from defined natural ecological habitats. However, it is also an appropriate term for the intra-species genetic erosion caused by domestication, post-domestication migration and, most recently, breeding of many of our major crops.
During the domestication of barley some 10,000 years ago (i.e. the conversion of wild germplasm into that suited to grow under cultivation), the proportion of available genetic diversity that made its way into the emerging domesticated gene pool was only a fraction of that present in the ‘founder’ wild populations. This so-called ‘bottleneck of domestication’ resulted in many potentially valuable versions of barley genes (called alleles) failing to make it into the new cultivated gene pool.
Then, as primitive agriculturalists migrated across the world, the crops they took with them were challenged to grow in a wide range of environments, facing many different biotic and abiotic threats. Responding to these challenges, through a number of processes, DNA in the barley genome lost detrimental and gained beneficial sequence variants which, combined with human selection forced the plant to adapt and ultimately flourish under specific conditions.
This process continued up until the last hundred or so years when plant breeding began to intensively select for specific changes in traits that increased productivity – potentially at the expense of those that confer resilience.
The novel strategy we are exploring tests whether we can efficiently retrieve valuable genetic biodiversity from wild barley species. By re-introducing ‘lost’ genetic biodiversity into the barley breeding gene pool, we believe there is an opportunity to increase resilience to our changing climate alongside associated environmental and regulatory issues (e.g. GHG emissions) while maintaining current levels of production.
Results
The most genetically diverse wild barley grows all over the near east fertile crescent (Israel, Lebanon, Iran, Iraq), a region considered both its centre of origin and diversity. We used variations in DNA sequences from over 1.5 million locations across the barley genome to characterise and structure the genetic diversity in a collection of 91 wild genotypes (individuals) from across this region (Figure 1). Using this information, we selected ten (Figure 1: yellow arrows) of the most diverse genotypes from across the geographical range to act as recipients of key ‘domestication genes’ from a popular contemporary barley cultivar (cv. ‘RGT Planet’) by traditional crossing. This is only possible because over the past 20 years we, and others in the barley genetics community, have identified and characterised barley domestication genes and genes conferring adaptation to specific mega-environments (e.g. the temperate Oceanic environment of western Europe) using modern genetics and genomics.
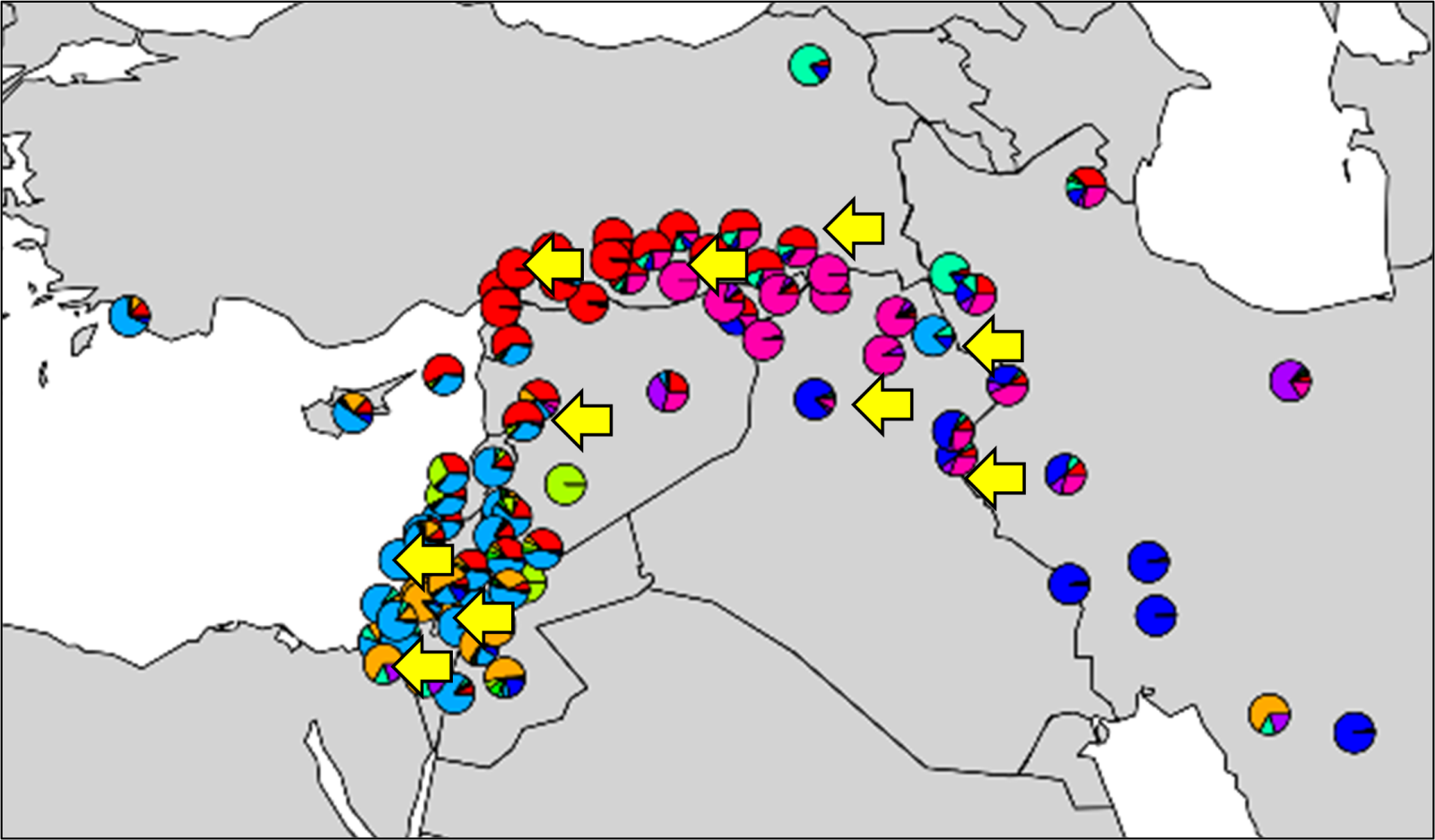
Figure 1. Geographical partitioning of diversity. The locations of 91 wild barley accessions colour coded based on diversity, are represented by longitude and latitude coordinates, clearly indicating geographic structuring based on variation. Yellow arrows indicate accessions chosen for R-Evolve to represent both genetic and geographical diversity (authors). Colours in each segment of each pie chart represent the proportion of the same ‘ancestral group’ in each individual.
The breeding scheme we used - and christened ‘R-evolve’ (for rapid evolution), and the potential of the wild germplasm is clearly illustrated as ‘hybrid vigor’, shown in Figure 2.

Figure 2. Hybrid vigor in an individual F2 plant (authors)
A key part of our process was the use of molecular diagnostics to identify plants containing desired combinations of domestications genes in the earliest possible generation (the so-called F2 generation). We only took forward the expected one-in-four offspring containing the domesticated version of a gene called BRITTLE RACHIS 1, which is responsible for retaining all of the grain on the spike (the barley inflorescence or ‘head’) at maturity. Having a non-brittle (i.e. tough) rachis is both necessary and sufficient to partially domesticate a wild barley plant. At the same time, we used molecular diagnostics for four other genes which help determine spring growth habit and plant height.
We then allowed selected individuals derived from each of the original ten plant families to self-fertilise for five more generations to generate a collection of 3700 genetically fixed (i.e. pure) ‘inbred lines’. To adhere to a highly constrained timescale we did this using a process that has been popularised by using the term speed breeding, where plants are subjected to artificial environmental conditions that accelerate plant development. This collection of 3700 inbred ‘R-evolved’ barley lines were then ready for phenotypic (trait) evaluation by exploring how they looked and performed in the field and polytunnel. Relevant preliminary evaluation data have been collected in summer 2024.
Next Steps
Our immediate priority is to analyse the phenotypic data derived from the inbred lines grown in the polytunnel and field to see whether any individuals perform (e.g. yield) as well, or potentially better, than the domesticated parent RGT Planet. We will evaluate their Genomic composition (wild vs. domesticated) using a so called ‘SNP genotyping platform’ that analyses variation at 50,000 DNA sequences from across the barley genome. Finally, in the original R-evolve strategy we planned to evaluate the inbred lines by making F1 hybrids with RGT Planet and evaluating their performance relative to the parents. In this approach, we evaluate whether a hybrids performance (as shown in Figure 2) is better than the mid parental value of each of the parents.
This approach only ‘works’ here because both parents are already effectively domesticated (i.e. comparisons to the original wild parent would be meaningless). If the F1 hybrids performance is better than the average value of the two parents then we conclude that the R-evolve wild parent contains versions of genes that are better than, or supplement, those in RGT Planet and could offer significant value as a parent for future breeding cycles. Ideally F1 hybrid performance should be tested under conditions matching future environmental and agronomic conditions.
Benefits
The global success of plant breeding has brought untold benefits to humankind, but, more than ever, we are facing environmental and regulatory challenges that could stall, or even reverse, the breeding progress achieved over the past 70 years.
Evidence suggests that part of the problem lies in the use of an increasingly narrow pool of genetic diversity in traditional crop breeding and as a result there is an urgent need to revitalise breeding gene pools by incorporating some of the natural genetic biodiversity that resides in the more than 7,000,000 crop plant genotypes, including wild species, held in gene banks across the globe.
Maintaining barley production in the face of demanding climatic shifts combined with broader environmental and regulatory challenges, will underpin vital food and drink end-use sectors, and support rural communities in generating significant value for the Scottish economy.
Project Partners
The James Hutton Institute
The University of Dundee
We partner with a complementary project funded by the Australian Research Council at the University of Adelaide (Prof Peter Langridge, Dr. Scott Boden and Dr. Suong Cu).